
Published datasets are available on my CV page. Here in PDF, copied from TABLE III.2 of my dissertation.
#Cite sequencher download
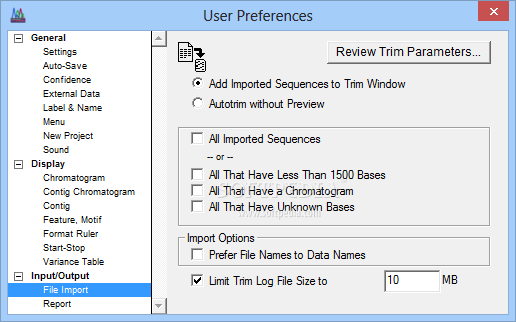
With standardized location and a complete list of all the samples (by collector number) I used them on, is available for download A list of all PCR and sequencing primers, I designed innumerable internal primers for this region, specific for my frogs. I also used mitochondrial DNA primers designed by Bob Macey forĪmplifying the "ND2-WANCY" region. Strabomantidae, depending on whom you ask.įor this work I designed original primers for PCR amplifying and sequencing a 1,300 base pair long portion of the c- myc gene,Ĭontaining intron 2 and portions of the flanking exons. (formerly Eleutherodactylus) in the family Brachycephalidae, or In myĬrawford (2003a) paper I cited this program as being available at which no longer exists due toįor my PhD thesis project I studied ecological genetics, phylogenetics, and molecular evolution in "dirt frogs" of the genus Craugastor Version 6.1.0 (not Sequencher!), a very handy DNA sequence analysis program written by Bailey Kessing, formerly of the
#Cite sequencher software
Software Software by Bailey KessingĪs a community service I am posting a copy of the program, Sequencer A plug for Brian O'Meara's old pageįor some concise tips on using PAUP*, check out Brian O'Meara's old web page. MULTIDIVTIME_HELPER and CONVERTER scriptsįrom the biokubuntu webpage. Prospective first-time users of multidivtime should also consider the following aids and alternatives: Updated version (for 2008) now provides tips on the closely related Thornian Time Travellerįirst time users should also consider BEAST as an alternative or a point of comparison. Step-by-step user's guide to multidistribute that will walk you through estbranches and multidivtime. The only catch is that this program is definitely
Polytomies or missing data are no problem. This program also allows (and encourages) simultaneous analysis of multiple loci. This program required no molecularĬlock, but the user should input some form of temporal constraints based on fossil data or a robust biogeographic hypothesis. Jeff Thorne and colleagues have designed a brilliant piece of software, multidivtime (aka, "multidistribute" or "Thornian Time Traveler"), for estimatingĭivergence times of samples on a given phylogeny based on DNA (or AA) sequences. Horribly detailed instructions to running multidivtime Molecular Phylogenetics and Evolution 47, 992-1004. Wang IJ, Crawford AJ, Bermingham E (2008) Phylogeography of the Pygmy Rain Frog ( Pristimantis ridens)Īcross the lowland tropical forests of Isthmian Central America. Same data set we originally applied our SOWH test protocol to: I was happy to see they tested their SOWHAT on the These authors now offer a cleverly named automated version,
My horribly detailed instructions to running a parametric bootstrap test. Of the subject, Anderson, Goldman and Rodrigo, available here. You probably should also consult the definative but not-quite-as-detailed SOWH guideline written by the masters Nowadays, I might recommend one try Mequite first. Of phylogenetic tree topology, aka, a SOWH test, downloadable as a RTF document. I wrote this horribly detailed instructions to running a parametric bootstrap test Horribly detailed instructions to running a SOWH test If you have any suggestions, comments, or questions, pleaseĭrop me a line at andrew ∀ dna.ac (replace ∀ with obviously). I have posted two informal but potentially useful documents I have written.īelow are some useful links as well. PhyLoTA: browse GenBank by phenetic clusters.Īlignment and tree building tools online. Tools for Evolutionary Genetic Analyses Quick links to databases and programs


 0 kommentar(er)
0 kommentar(er)
